1 Introduction
The Model Description Language (MDL) and the Pharmacometrics Markup Language standard (PharmML) (M. Swat et al. 2015) have been developed to convey information about pharmacometrics models and tasks. The goal of each language is to do this consistently between modellers (using MDL) and between software target tools (using PharmML).
MDL is a human writeable and human readable language designed to describe pharmacometric models. It is intended to be largely agnostic about the choice of target tool. MDL should facilitate clear and unambiguous definition of models, with information conveyed in a consistent manner to the PharmML representation and onwards to the target software specific code.
An important concept in the MDL is the separation of data, parameter, model and task descriptions into independent objects rather than combining these in a single file (such as in NONMEM (Bauer 2011)). This supports reuse and interchange of the objects which define each component of the model and related modelling task. This independence means that model objects stored in the DDMoRe Model Repository may be combined with user objects outside the repository e.g. a Model Object, Parameter Object and a {Task Properties Object} may be taken from the Repository and combined with user defined Data Object. This may be useful when a user wishes to assess whether a library model is predictive for their dataset, as a preliminary step before further model refinement.
These facets (target software agnostic code + independence of MDL objects) mean that model definition using MDL is more verbose than code written specifically for any specific target tool. However the principle concept of MDL is that model code is written once and used in many different tools. For estimation, simulation, optimal design. So time spent writing code initially is saved in the longer term since MDL eliminates the need to recode models for different tasks and different software tools.
1.1 Why write a new language?
A key deliverable of the DDMoRe project is a unified Model Description Language (MDL), based on established principles, designed to be easily read and written. It is designed to facilitate easy uptake by modellers already experienced in other model definition languages, and will allow the definition of any model-based analysis.
Several languages have been created to support M&S activities. Examples of widely used languages are NONMEM (NMTRAN), Monolix (MLXTRAN) (Lavielle 2012), BUGS (Lunn et al. 2000) and MATLAB (MATLAB 2000). However, none are shared, creating difficulties for comparison and integration. Many tools have overlapping functionality, and so the choice of one tool over another is driven largely by user preference, availability of tools, experience of the analyst and whether there is sufficient experience readily available to the analyst to provide support and advice on model building techniques specific for the tool in question. Considerable effort is currently required when moving the model from one software tool to another, as models always have to be recoded in the target software tool language, by hand. A significant need exists to rectify this situation, which DDMoRe is addressing.
Another common situation is using models which were developed by a third party using software that we do not have available. In that case we must try to re-encode the model before we can start using it or developing it further. This can be difficult because we need to ensure that we have all the information to construct the model in a different language. Do we have all the necessary files, settings, subroutines, functions available to us? Are the assumptions used in the model adequately annotated or described in supporting documentation? Do we have understanding of any tool-specific tricks and techniques that allow the model to work in the original software?
MDL provides a user interface to describe models using a common language standard. The aim is that the user writes the model (Model Object) once, in MDL, then uses this model in the tools they require (and have available) in order to complete their M&S tasks, without any tool specific recoding. This interoperability is a core deliverable of the DDMoRe project. (Harnisch et al. 2013)
Additionally, MDL is intended as a standard for communication of models. An analyst who only uses one tool may wish to convey their model to a third party. MDL provides the means to describe the model in a way that is consistent and provides complete information about the model (without any reference to target tool specific code). MDL is designed to focus on describing WHAT the model conveys, rather than focussing on the HOW of implementation. This has an impact on the structure and features used in MDL, but it should aid clarity and reduce ambiguity.
1.2 Integrated language standards
As described above, MDL provides the user focused layer of model description. This facilitates user understanding and model sharing between analysts.
PharmML provides the software interchange standard within DDMoRe to facilitate the transfer of models between target tools by ensuring that all of the necessary information about the model is captured and can be translated automatically to any given target tool that has an appropriate PharmML converter.
ProbOnto (M. J. Swat, Grenon, and Wimalaratne 2016) is an ontology and knowledge base that has been developed to describe probability distributions in a consistent and unambiguous way, as well as defining their functions, characteristics and the relationships between distributions.
The Standard Output object (SO) standard provides a consistent format for M & S results and outputs. Its availability as an object within R provides interchange and integration between existing R packages for M & S tasks within the DDMoRe infrastructure.
MDL, PharmML and the SO are the basis for interoperability which is one of the core deliverables of the DDMoRe project.
1.3 MDL in use
Very few models can be retrieved from a repository or library, be fit to any given set of data and pronounced valid for inference without further assessment or changes. Thus, the process of fitting models to data, assessing the fit through model diagnostics is an iterative process, culminating in selecting the model which is parsimonious and fit for its purpose in the inferential step decision making, making predictions for future populations of interest, selecting dose or dosage regimen etc. The combination of features in MDL and the ddmore R package facilitates that process. MDL’s structure makes changes to data, models, parameters, tasks transparent making it clear exactly which elements are changing and which are constant across steps. Using an R script to define M & S task workflow facilitates an unbroken workflow for a given model and dataset, from exploratory analysis, estimation, diagnostics and simulation across a variety of tools without having to recode the model.
1.4 MDL components and structure
The MDL objects are typically defined in a file with extension .mdl. Models may also be stored and retrieved from the DDMoRe Repository either as MDL or PharmML. The key concept in MDL is that these objects can be passed to any target software for use in modelling tasks: estimation, model diagnostics and evaluation, simulation, optimal design.
An overview of the currently specified MDL objects is shown in Figure 1.1.
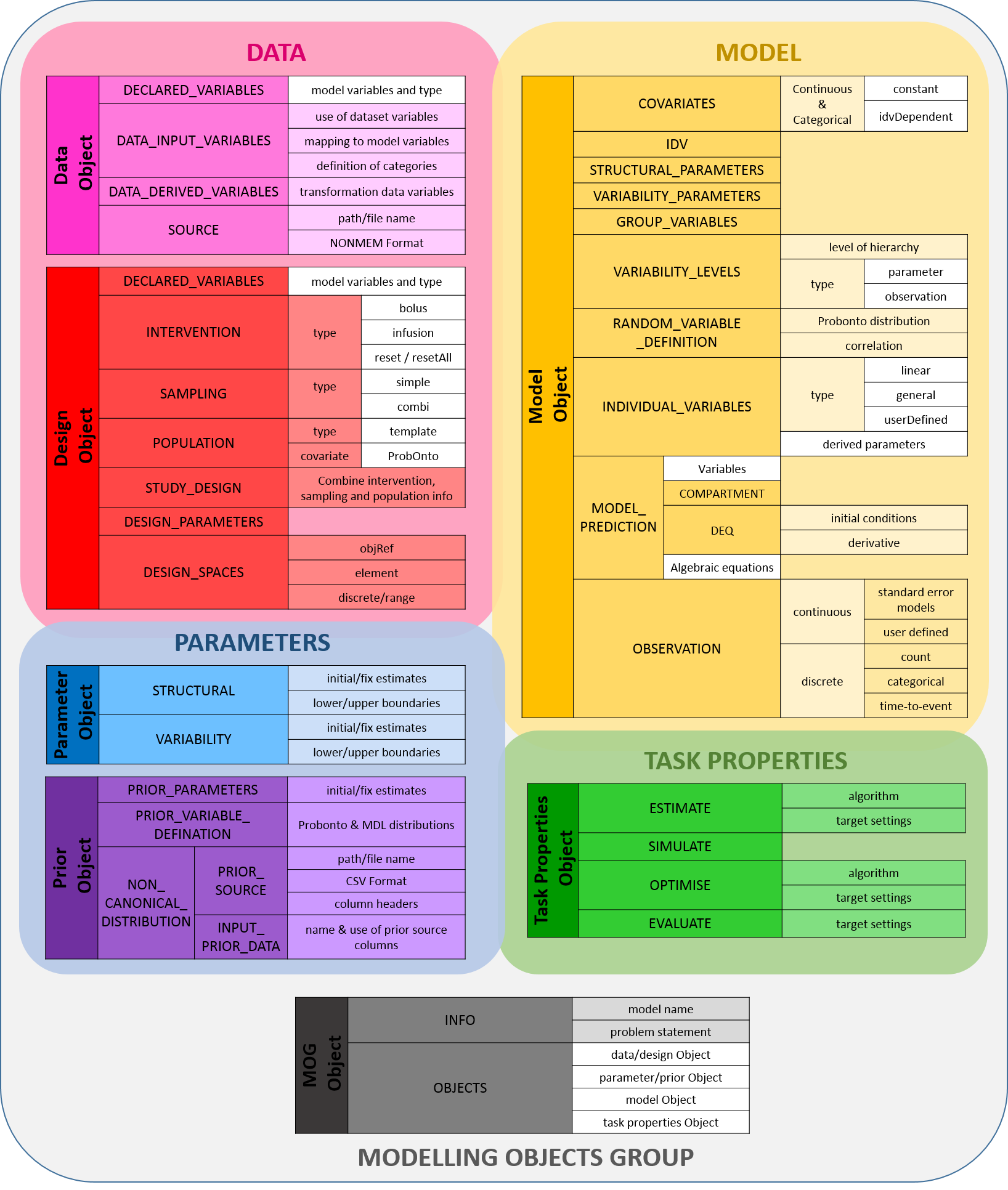
Figure 1.1: MDL Objects
The MDL is used to specify the inputs and the model used in an M & S task. It does this with four MDL objects defining the model, parameters, data and task properties. An additional object is used to specify the group of objects required for a given task which is known as the Modelling Objects Group (MOG).
The Model Object is the core element of the MDL and Modelling Objects Group (MOG). It defines the mathematical and statistical properties of the model by defining the structural, covariate, variability and observation models. While other objects may change depending on task, the Model Object will typically be unchanged for tasks associated with that model e.g. data visualisation, parameter estimation, model diagnostics, prediction, simulation or optimal design.
The Data Object describes the source of the data and the attributes of each of the data variables. It allows the user to define the inputs to the model and how these inputs and observed data are to be used in definition of the model. It may also be used with data visualisation tools without a Model Object.
The Design Object defines the design parameters interventions, sampling schedules, covariate distributions, populations, study arms - for optimal design or design evaluation, and also for simulation. The Design Object replaces the Data Object in these cases.
The Parameter Object provides values for structural and variability parameters, including bounds on the parameter values for use in estimation. These can be fixed or initial values with associated constraints for parameter estimation or an instantiation of model parameters for use in making predictions or simulations.
The Prior Object defines prior distributions and values of the parameters when performing Bayesian estimation of parameters. It replaces the Parameter Object in this case.
The Task Properties Object contains settings specific to the task which will be passed on to the target software e.g. when estimating parameters it will define the estimation algorithm and associated settings for the algorithm.
It is through combination of the Model Object with other objects that we instantiate the model - linking inputs and observations from the Data Object or Design Object, parameter values from the Parameter Object or Design Object and information about the task settings in the Task Properties Object with the Model Object to form a Modelling Objects Group (MOG) ready for executing a modelling, simulation or optimal design task.
Objects are defined and stored in a MDL file with extension .mdl. It is possible to define more than one Model, Data, Design, Parameter Prior and Task Properties Object within a single MDL file. The MOG Object defines specific individual objects within an MDL file for a given task. Most commonly there will be one object of each type of information in a MDL file used for a task.
1.4.1 Independence of MDL objects
Typically, existing software has used control files that bring the elements in MDL (data, parameters, model and task definitions) together in control, model or project file(s). What is new in the MDL is the concept that the elements of the Modelling Object Group (data, parameters, model, task properties) are distinct and independent, allowing the user to combine new data and parameters applicable to their situation with an existing model. Within a M&S task workflow it is easy to see how the core Model Object remains unchanged between estimating parameters, performing model diagnostics, making predictions and simulation future outcomes. The fact that the elements of the MDL are exchangeable also makes it easier to see exactly what elements change between these M & S workflow steps.
Independence of objects also means that the Model Object should be independent of the data and, as a consequence, more easy to read and interpret without needing to have the data to hand. Having an independent Task Properties Object means that the user may store their preferred settings for tasks and target software, suitable for reuse across models and modelling tasks, to facilitate comparison between target software and to ensure reproducibility of results.
Independence of the MDL objects entails defining the contents of each object in isolation. Variables from another MDL object must be declared in the object in which they are referred to e.g. if we need to refer to the Model Object OBSERVATION block variable Y in the Data Object DATA_INPUT_VARIABLES block then we must declare a matching variable Y in the Data Object.
1.5 Task Execution with the ddmore R package
To perform tasks with the model, the user will need to use the ddmore R package. This package contains functions for executing commonly used tasks on the MDL file. The R functions can read and parse MDL objects from a MDL file to create R object representations of MDL, which can then be manipulated within R. These representations of the MDL objects can be combined to form a MOG and then written back to a new MDL file. This means that an MDL file can contain the core Model Object and associated Data, Design, Parameter, Prior and multiple Task Properties Objects which can then be combined into MOGs ready to perform specific tasks. This aids reproducibility since with one MDL file, data set and associated R script the user can perform multiple steps in a pharmacometric workflow for a given model.
Estimation using the estimate function takes as input the user specified MDL file or a MOG Object defined within R. Additional functions allow the user to call modelling tools such as Perl speaks NONMEM (PsN) [8]. Each task produces a Standard Output (SO) object in R which may be the final output or used in subsequent tasks using functions from the ddmore R package or other R packages and commands.
Using R as the language for defining the workflow for M & S tasks with MDL objects allows analysts to tap into existing R packages for performing those tasks. The ddmore package R functions are provided to read and extract information from the SO object and to convert between this, Xpose (Jonsson and Karlsson 1998), mlxR (M. Lavielle, n.d.), PFIM (Bazzoli, Retout, and Mentré 2010) and PopED (Foracchia et al. 2004)R packages. The ddmore R package will extend and enhance what the analyst can do with existing R packages through the common standards that the DDMoRe project brings.
Task properties and settings defined in the Task Properties Object of MDL are distinct from arguments to the R functions for executing tasks. The Task Properties Object provides information to the appropriate target software about the particular settings and options required for a given task. The R function arguments are command line settings or options which are employed when invoking the target software. The Task Properties Object may define the estimation algorithm and associated settings for NONMEM, but the command line options for PsN provided from the ddmore functions govern how NONMEM should be called by PsN. For example, Task Properties specifies an ESTIMATION block with estimation method set to FOCEI, while the arguments of the bootstrap.PsN function in R allow the user to set bootstrap options from PsN such as threads, stratify_on etc. (See PsN bootstrap documentation for more details on PsN bootstrap options).
1.6 The MDL Integrated Development Environment
The MDL Integrated Development Environment (MDL-IDE) is a software platform for writing models with MDL. The MDL editor within the MDL-IDE implements the rules of the language through recognising MDL constructs and having a defined grammar and it ensures that MDL models are syntactically correct and result in valid PharmML. The MDL-IDE also provides additional tools giving access to an R editor and console – so that the user can not only develop models, but execute tasks with them and define task workflow through R scripts.
The MDL-IDE gives warnings when the user writes MDL that will result in valid PharmML, but where MDL constructs are used that may not be interoperable. It gives errors when the user writes code that breaks MDL grammar rules and that will result in invalid PharmML.
1.7 On interoperability
A key goal of the DDMoRe project is to have an intoperability framework in which models are written in a consistent language, translated to PharmML and from there converted to target software code. Before the DDMoRe project no existing language standard existed across target software used in pharmacometrics modelling, and while the underlying models could be expressed consistently in mathematical and statistical terms, the implementation of any given model varied by tool and by user according to their experience with a given target software tool.
There is some flexibility within MDL around how the user can express the mathematical and statistical models. Having flexibility allows the user to encode models quickly in a common language (MDL) which can then be shared with others and mutually understood. This flexibility also facilitates encoding in a given target when that language construct does not have a parallel in other tools. However, we STRONGLY encourage the user to encode the majority of models in a way that will facilitate interoperability. There are MDL constructs that facilitate interoperability these generally appear as built-in functions which translate to specific constructs in PharmML and the target software. These constructs cover many typical models and are designed to allow the user to generate code quickly and have high confidence that it will be interoperable across tools.
The Model Description Language Interactive Development Environment (MDL-IDE) should assist the user in ensuring that the models encoded are valid MDL (and as a consequence, also valid PharmML). Not all models will result in code which can be readily converted to all target tools.
These interoperability constructs will be highlighted in the subsequent sections, but users should pay particular attention to sections on the use of GROUP_VARIABLES,INDIVIDUAL_VARIABLES and the MODEL_PREDICTION.
1.8 Evolution of MDL
Development of MDL has been led and influenced by domain experts in M & S, computer language development, system interchange language development (markup languages), and developers of software systems. In developing MDL we have looked at features in established M&S languages, as mentioned above, and aimed to pick out features that will facilitate interoperability, while retaining the flexibility in these languages to describe complex models. The current MDL implementation focusses on interoperability in order to demonstrate that capability. The language standards in MDL, PharmML and SO are the key to eliminating the recoding necessary to pass models between tools used for different M&S tasks.
The MDL language attempts to balance consistency and clarity in definitions, with interoperability and flexibility in translation to PharmML and on to target software. It will continue to evolve to incorporate new features, extending the range of models that can be expressed using the language.
Trying to define a language that maps to all possible models as defined in all possible tools is virtually impossible. However, having a well-defined software interchange standard (PharmML) and mapping MDL into PharmML allows us to focus on describing model features with one target in mind PharmML. The two languages MDL and PharmML - have evolved during the course of the project. The aim is that these two languages should go hand in hand that MDL should convey in an accessible, user (analyst) friendly way, the models that can be encoded in PharmML.
Converter tools then interpret the PharmML rather than the MDL for each software target. Testing this conversion and comparing output downstream allows us to check that the translation results in comparable models.
Future pharmacometrics tools could provide converters to import and export PharmML or use MDL directly as the model specification language. It is our hope that the DDMoRe standards would facilitate more consistency, better understanding of models as well as interoperability between modelling and simulation tools in the future.
References
Swat, MJ, S Moodie, SM Wimalaratne, NR Kristensen, M Lavielle, A Mari, P Magni, et al. 2015. “Pharmacometrics Markup Language (PharmML): Opening New Perspectives for Model Exchange in Drug Development.” CPT: Pharmacometrics & Systems Pharmacology 4 (6). Wiley-Blackwell: 316–19. doi:10.1002/psp4.57.
Bauer, R. 2011. NONMEM Users Guide Introduction to Nonmem 7.2. 0. ICON Development Solutions Ellicott City, MD. Ellicott City, MD: Icon Development Solutions.
Lavielle, M. 2012. MONOLIX User Guide. Lixoft, Orsay, France. Antony, France; Inria, Orsay, France: Monolix, Lixoft. http://www.lixoft.eu/.
Lunn, David J., Andrew Thomas, Nicky Best, and David Spiegelhalter. 2000. “WinBUGS - a Bayesian Modelling Framework: Concepts, Structure, and Extensibility.” Statistics and Computing 10 (4). Springer Nature: 325–37. doi:10.1023/a:1008929526011.
MATLAB. 2000. Natick, MA: The MathWorks Inc. http://nl.mathworks.com/products/matlab.
Harnisch, L, I Matthews, J Chard, and M O Karlsson. 2013. “Drug and Disease Model Resources: A Consortium to Create Standards and Tools to Enhance Model-Based Drug Development.” CPT: Pharmacometrics & Systems Pharmacology 2 (3). Wiley-Blackwell: e34. doi:10.1038/psp.2013.10.
Swat, Maciej J., Pierre Grenon, and Sarala Wimalaratne. 2016. “ProbOnto: Ontology and Knowledge Base of Probability Distributions.” Bioinformatics 32 (17). Oxford University Press (OUP): 2719–21. doi:10.1093/bioinformatics/btw170.
Jonsson, E.Niclas, and Mats O Karlsson. 1998. “Xposean S-PLUS Based Population Pharmacokinetic/Pharmacodynamic Model Building Aid for NONMEM.” Computer Methods and Programs in Biomedicine 58 (1). Elsevier BV: 51–64. doi:10.1016/s0169-2607(98)00067-4.
Bazzoli, Caroline, Sylvie Retout, and France Mentré. 2010. “Design Evaluation and Optimisation in Multiple Response Nonlinear Mixed Effect Models: PFIM 3.0.” Computer Methods and Programs in Biomedicine 98 (1). Elsevier BV: 55–65. doi:10.1016/j.cmpb.2009.09.012.
Foracchia, Marco, Andrew Hooker, Paolo Vicini, and Alfredo Ruggeri. 2004. “Poped, a Software for Optimal Experiment Design in Population Kinetics.” Computer Methods and Programs in Biomedicine 74 (1). Elsevier BV: 29–46. doi:10.1016/s0169-2607(03)00073-7.